Ocean reanalyses in 2010 (Deepwater Horizon year)¶
[1]:
bathy_URL = 'http://geoport.whoi.edu/thredds/dodsC/bathy/etopo1_bed_g2'
glb90_URL = 'http://tds.hycom.org/thredds/dodsC/GLBa0.08/expt_90.8'
glb19_URL = 'http://tds.hycom.org/thredds/dodsC/GLBu0.08/expt_19.1/2010'
gom31_URL = 'http://tds.hycom.org/thredds/dodsC/GOMl0.04/expt_31.0/2010'
gom50_URL = 'http://tds.hycom.org/thredds/dodsC/GOMu0.04/expt_50.1/data/netcdf/2010'
glb53_URL = 'http://tds.hycom.org/thredds/dodsC/GLBv0.08/expt_53.X/data/2010'
[3]:
import xarray as xr
%pylab inline
Populating the interactive namespace from numpy and matplotlib
[4]:
# test opendap connection
xr.open_dataset(bathy_URL)
[4]:
<xarray.Dataset>
Dimensions: (lat: 10801, lon: 21601)
Coordinates:
* lon (lon) float64 -180.0 -180.0 -180.0 -179.9 ... 180.0 180.0 180.0
* lat (lat) float32 -90.0 -89.98333 -89.96667 ... 89.96667 89.98333 90.0
Data variables:
topo (lat, lon) float32 ...
Attributes:
Conventions: COARDS/CF-1.0
title: NGDC ETOPO1 Global DEM (1 min)
history: grdreformat ETOPO1_Bed_g.int.grd=ni ETOPO1_Bed_g.grd=ns
GMT_version: 4.3.1b
node_offset: 0
[5]:
glb90 = xr.open_dataset(glb90_URL, decode_times=False)
[6]:
glb90
[6]:
<xarray.Dataset>
Dimensions: (Depth: 33, MT: 606, X: 4500, Y: 3298)
Coordinates:
* Y (Y) int32 1 2 3 4 5 ... 3294 3295 3296 3297 3298
* X (X) int32 1 2 3 4 5 ... 4496 4497 4498 4499 4500
* Depth (Depth) float32 0.0 10.0 20.0 ... 5000.0 5500.0
* MT (MT) float64 3.957e+04 3.958e+04 ... 4.018e+04
Date (MT) float64 ...
Latitude (Y, X) float32 ...
Longitude (Y, X) float32 ...
Data variables:
mld (MT, Y, X) float32 ...
mlp (MT, Y, X) float32 ...
qtot (MT, Y, X) float32 ...
emp (MT, Y, X) float32 ...
surface_temperature_trend (MT, Y, X) float32 ...
surface_salinity_trend (MT, Y, X) float32 ...
ssh (MT, Y, X) float32 ...
salinity (MT, Depth, Y, X) float32 ...
temperature (MT, Depth, Y, X) float32 ...
u (MT, Depth, Y, X) float32 ...
v (MT, Depth, Y, X) float32 ...
Attributes:
Conventions: CF-1.0
title: HYCOM GLBa0.08
institution: Naval Research Laboratory
source: HYCOM archive file
experiment: 90.8
history: archv2ncdf2d
[7]:
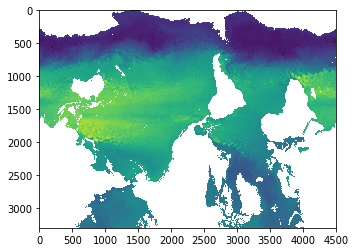
ssh = glb90.ssh
imshow(ssh[0,:,:])
[7]:
<matplotlib.image.AxesImage at 0x318326dd8>
[8]:
gom50 = xr.open_dataset(gom50_URL, decode_times=False)
[9]:
gom50
[9]:
<xarray.Dataset>
Dimensions: (depth: 40, lat: 346, lon: 541, time: 2920)
Coordinates:
* depth (depth) float64 0.0 2.0 4.0 6.0 ... 2.5e+03 3e+03 4e+03 5e+03
* lat (lat) float64 18.12 18.16 18.2 18.24 ... 31.8 31.84 31.88 31.92
* lon (lon) float64 -98.0 -97.96 -97.92 -97.88 ... -76.48 -76.44 -76.4
* time (time) float64 8.767e+04 8.768e+04 ... 9.643e+04 9.643e+04
Data variables:
tau (time) float64 ...
water_u (time, depth, lat, lon) float32 ...
water_v (time, depth, lat, lon) float32 ...
water_temp (time, depth, lat, lon) float32 ...
salinity (time, depth, lat, lon) float32 ...
surf_el (time, lat, lon) float32 ...
Attributes:
classification_level: UNCLASSIFIED
distribution_statement: Approved for public release. Distribution unli...
downgrade_date: not applicable
classification_authority: not applicable
institution: Naval Oceanographic Office
source: HYCOM archive file
history: archv2ncdf3z
field_type: instantaneous
Conventions: CF-1.6 NAVO_netcdf_v1.1
[10]:
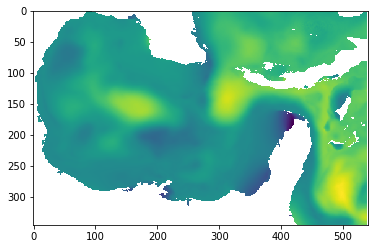
ssh50 = gom50.surf_el
imshow(ssh50[0,:,:])
[10]:
<matplotlib.image.AxesImage at 0x32accf0f0>
[ ]:
