Note
Click here to download the full example code
Isentropic Analysis¶
The MetPy function metpy.calc.isentropic_interpolation allows for isentropic analysis from model analysis data in isobaric coordinates.
from datetime import datetime, timedelta
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
import metpy.calc
from metpy.units import units
from netCDF4 import num2date
import numpy as np
from siphon.catalog import TDSCatalog
Getting the data
In this example, the latest GFS forecasts data from the National Centers for Environmental Information (https://nomads.ncdc.noaa.gov) will be used, courtesy of the Univeristy Corporation for Atmospheric Research Thredds Data Server.
# Latest GFS Dataset
cat = TDSCatalog('http://thredds-jetstream.unidata.ucar.edu/thredds/catalog/grib/'
'NCEP/GFS/Global_0p5deg/catalog.xml')
ncss = cat.latest.subset()
# Find the start of the model run and define time range
start_time = ncss.metadata.time_span['begin']
start = datetime.strptime(start_time, '%Y-%m-%dT%H:%M:%Sz')
end = start + timedelta(hours=9)
# Query for Latest GFS Run
gfsdata = ncss.query().time_range(start, end).accept('netcdf4')
gfsdata.variables('Temperature_isobaric',
'u-component_of_wind_isobaric',
'v-component_of_wind_isobaric',
'Relative_humidity_isobaric').add_lonlat()
# Set the lat/lon box for the data you want to pull in.
# lonlat_box(north_lat,south_lat,east_lon,west_lon)
gfsdata.lonlat_box(-150, -50, 15, 65)
# Actually getting the data
data = ncss.get_data(gfsdata)
dtime = data.variables['Temperature_isobaric'].dimensions[0]
dlev_hght = data.variables['Temperature_isobaric'].dimensions[1]
dlev_uwnd = data.variables['u-component_of_wind_isobaric'].dimensions[1]
lat = data.variables['lat'][:]
lon = data.variables['lon'][:]
lev_hght = data.variables[dlev_hght][:] * units.Pa
lev_uwnd = data.variables[dlev_uwnd][:] * units.Pa
# Due to a different number of vertical levels find where they are common
_, _, common_ind = np.intersect1d(lev_uwnd, lev_hght, return_indices=True)
times = data.variables[dtime]
vtimes = num2date(times[:], times.units)
temps = data.variables['Temperature_isobaric']
tmp = temps[:, common_ind, :, :] * units.kelvin
uwnd = data.variables['u-component_of_wind_isobaric'][:] * units.meter / units.second
vwnd = data.variables['v-component_of_wind_isobaric'][:] * units.meter / units.second
relh = data.variables['Relative_humidity_isobaric'][:]
To properly interpolate to isentropic coordinates, the function must know the desired output isentropic levels. An array with these levels will be created below.
isentlevs = np.arange(310, 316, 5) * units.kelvin
Conversion to Isentropic Coordinates
Once model data in isobaric coordinates has been pulled and the desired isentropic levels created, the conversion to isentropic coordinates can begin. Data will be passed to the function as below. The function requires that isentropic levels, isobaric levels, and temperature be input. Any additional inputs (in this case relative humidity, u, and v wind components) will be linearly interpolated to isentropic space.
isent_anal = metpy.calc.isentropic_interpolation(isentlevs, lev_uwnd, tmp,
relh, uwnd, vwnd, axis=1)
The output is a list, so now we will separate the variables to different names before plotting.
isentprs, isentrh, isentu, isentv = isent_anal
A quick look at the shape of these variables will show that the data is now in isentropic coordinates, with the number of vertical levels as specified above.
print(isentprs.shape)
print(isentrh.shape)
print(isentu.shape)
print(isentv.shape)
Out:
(4, 2, 101, 201)
(4, 2, 101, 201)
(4, 2, 101, 201)
(4, 2, 101, 201)
Plotting the Isentropic Analysis
Set up our projection
crs = ccrs.LambertConformal(central_longitude=-100.0, central_latitude=45.0)
# Set up our array of latitude and longitude values and transform to
# the desired projection.
clons, clats = np.meshgrid(lon, lat)
# Get data to plot state and province boundaries
states_provinces = cfeature.NaturalEarthFeature(
category='cultural',
name='admin_1_states_provinces_lakes',
scale='50m',
facecolor='none')
level = 0
FH = 0
fig = plt.figure(1, figsize=(14., 12.))
ax = plt.subplot(111, projection=crs)
# Set plot extent
ax.set_extent((-121., -74., 25., 50.), crs=ccrs.PlateCarree())
ax.coastlines('50m', edgecolor='black', linewidth=0.75)
ax.add_feature(states_provinces, edgecolor='black', linewidth=0.5)
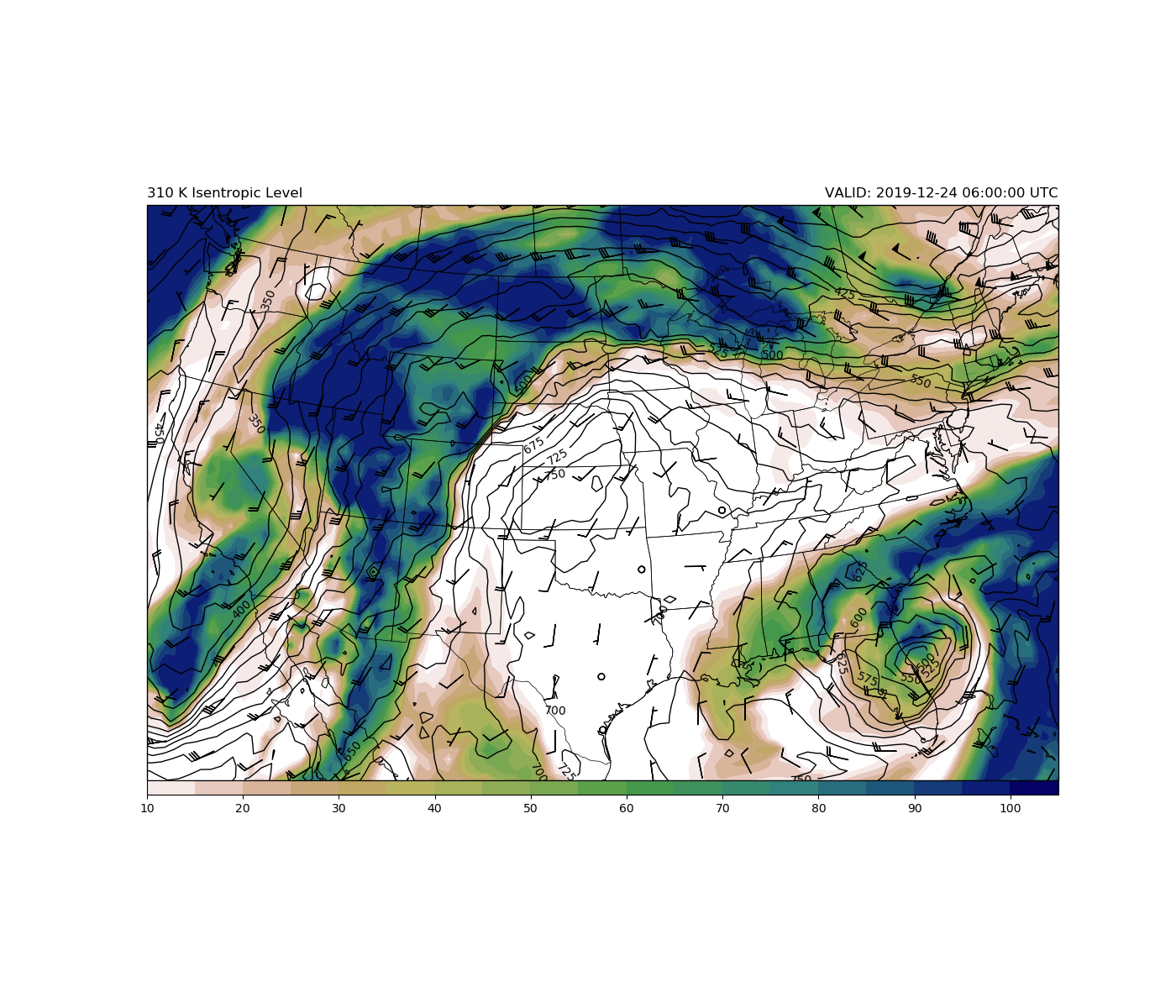
# Plot the 300K surface
clevisent = np.arange(0, 1000, 25)
cs = ax.contour(clons, clats, isentprs[FH, level, :, :], clevisent,
transform=ccrs.PlateCarree(),
colors='k', linewidths=1.0, linestyles='solid')
plt.clabel(cs, fontsize=10, inline=1, inline_spacing=7,
fmt='%i', rightside_up=True, use_clabeltext=True)
cf = ax.contourf(clons, clats, isentrh[FH, level, :, :], range(10, 106, 5),
transform=ccrs.PlateCarree(),
cmap=plt.cm.gist_earth_r)
plt.colorbar(cf, orientation='horizontal', extend=max, aspect=65, pad=0,
extendrect='True')
wind_slice = [FH, level, slice(None, None, 5), slice(None, None, 5)]
ax.barbs(clons[wind_slice[2:]], clats[wind_slice[2:]],
isentu[wind_slice].m, isentv[wind_slice].m, length=6,
transform=ccrs.PlateCarree())
# Make some titles
plt.title('{:.0f} K Isentropic Level'.format(isentlevs[level].m), loc='left')
plt.title('VALID: {:s} UTC'.format(str(vtimes[FH])), loc='right')
plt.show()
Out:
/home/travis/miniconda/envs/gallery/lib/python3.7/site-packages/numpy/ma/core.py:3172: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
/home/travis/miniconda/envs/gallery/lib/python3.7/site-packages/numpy/ma/core.py:3204: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
mout = _mask[indx]
Total running time of the script: ( 0 minutes 41.007 seconds)