Note
Click here to download the full example code
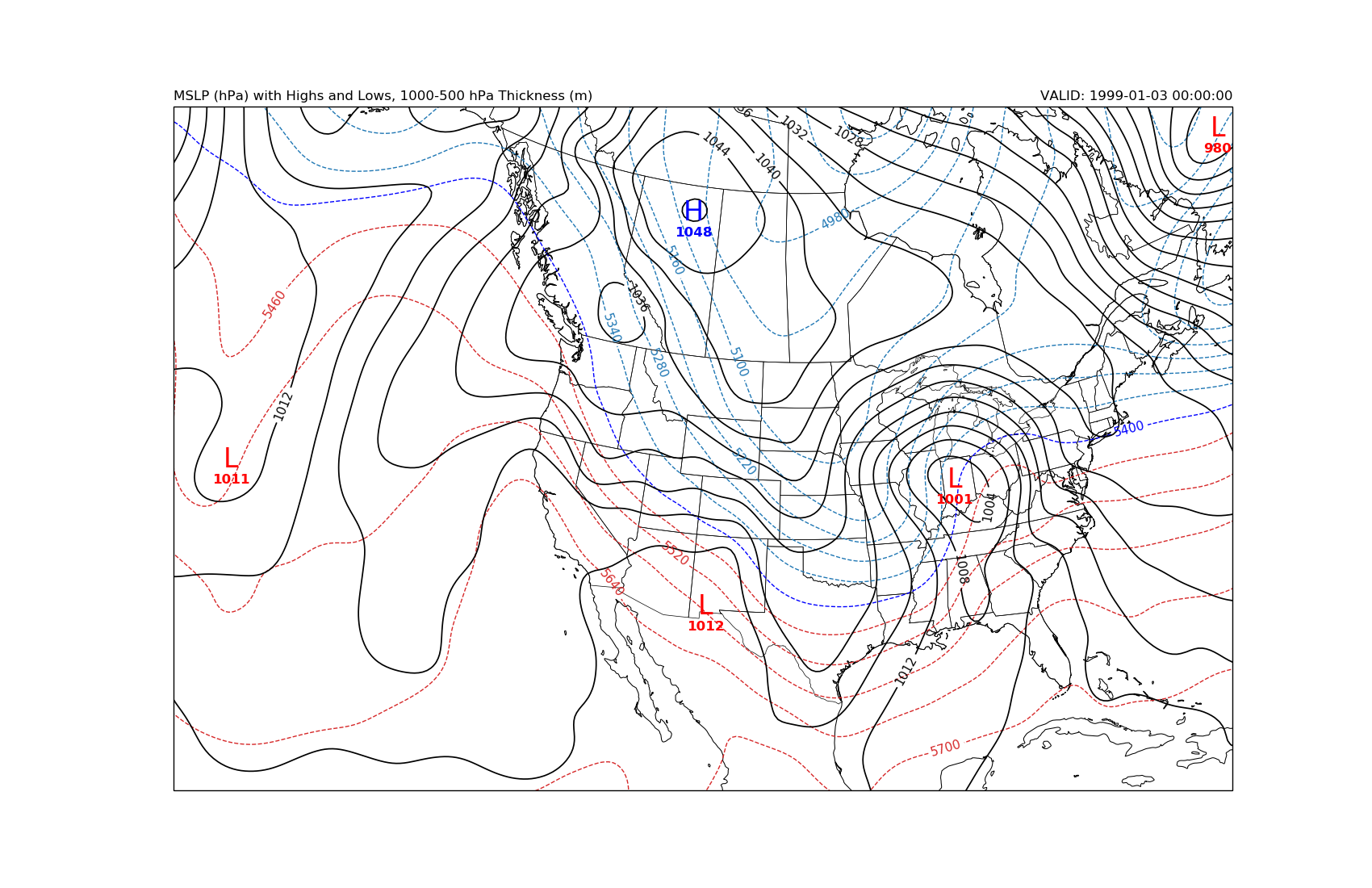
MSLP and 1000-500 hPa Thickness with High and Low Symbols¶
Plot MSLP, calculate and plot 1000-500 hPa thickness, and plot H and L markers. Beyond just plotting a few variables, in the example we use functionality from the scipy module to find local maximum and minimimum values within the MSLP field in order to plot symbols at those locations.
Imports
from datetime import datetime
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
from metpy.units import units
from netCDF4 import num2date
import numpy as np
from scipy.ndimage import gaussian_filter
from siphon.ncss import NCSS
Function for finding and plotting max/min points
def plot_maxmin_points(lon, lat, data, extrema, nsize, symbol, color='k',
plotValue=True, transform=None):
"""
This function will find and plot relative maximum and minimum for a 2D grid. The function
can be used to plot an H for maximum values (e.g., High pressure) and an L for minimum
values (e.g., low pressue). It is best to used filetered data to obtain a synoptic scale
max/min value. The symbol text can be set to a string value and optionally the color of the
symbol and any plotted value can be set with the parameter color
lon = plotting longitude values (2D)
lat = plotting latitude values (2D)
data = 2D data that you wish to plot the max/min symbol placement
extrema = Either a value of max for Maximum Values or min for Minimum Values
nsize = Size of the grid box to filter the max and min values to plot a reasonable number
symbol = String to be placed at location of max/min value
color = String matplotlib colorname to plot the symbol (and numerica value, if plotted)
plot_value = Boolean (True/False) of whether to plot the numeric value of max/min point
The max/min symbol will be plotted on the current axes within the bounding frame
(e.g., clip_on=True)
"""
from scipy.ndimage.filters import maximum_filter, minimum_filter
if (extrema == 'max'):
data_ext = maximum_filter(data, nsize, mode='nearest')
elif (extrema == 'min'):
data_ext = minimum_filter(data, nsize, mode='nearest')
else:
raise ValueError('Value for hilo must be either max or min')
mxy, mxx = np.where(data_ext == data)
for i in range(len(mxy)):
ax.text(lon[mxy[i], mxx[i]], lat[mxy[i], mxx[i]], symbol, color=color, size=24,
clip_on=True, horizontalalignment='center', verticalalignment='center',
transform=transform)
ax.text(lon[mxy[i], mxx[i]], lat[mxy[i], mxx[i]],
'\n' + str(np.int(data[mxy[i], mxx[i]])),
color=color, size=12, clip_on=True, fontweight='bold',
horizontalalignment='center', verticalalignment='top', transform=transform)
Get NARR data
dattim = datetime(1999, 1, 3, 0)
ncss = NCSS('https://www.ncei.noaa.gov/thredds/ncss/grid/narr-a-files/{0:%Y%m}/{0:%Y%m%d}/'
'narr-a_221_{0:%Y%m%d}_{0:%H}00_000.grb'.format(dattim))
query = ncss.query()
query.all_times().variables('Pressure_reduced_to_MSL_msl',
'Geopotential_height_isobaric').add_lonlat().accept('netcdf')
data = ncss.get_data(query)
Extract data into variables
# Grab pressure levels
plev = list(data.variables['isobaric1'][:])
# Grab lat/lons and make all lons 0-360
lats = data.variables['lat'][:]
lons = data.variables['lon'][:]
lons[lons < 0] = 360 + lons[lons < 0]
# Grab valid time and get into datetime format
time = data['time2']
vtime = num2date(time[:], units=time.units)
# Grab MSLP and smooth, use MetPy Units module for conversion
EMSL = data.variables['Pressure_reduced_to_MSL_msl'][:] * units.Pa
EMSL.ito('hPa')
mslp = gaussian_filter(EMSL[0], sigma=3.0)
# Grab pressure level data
hght_1000 = data.variables['Geopotential_height_isobaric'][0, plev.index(1000)]
hght_500 = data.variables['Geopotential_height_isobaric'][0, plev.index(500)]
# Calculate and smooth 1000-500 hPa thickness
thickness_1000_500 = gaussian_filter(hght_500 - hght_1000, sigma=3.0)
Set map and data projections for use in mapping
# Set projection of map display
mapproj = ccrs.LambertConformal(central_latitude=45., central_longitude=-100.)
# Set projection of data
dataproj = ccrs.PlateCarree()
# Grab data for plotting state boundaries
states_provinces = cfeature.NaturalEarthFeature(
category='cultural',
name='admin_1_states_provinces_lakes',
scale='50m',
facecolor='none')
Create figure and plot data
fig = plt.figure(1, figsize=(17., 11.))
ax = plt.subplot(111, projection=mapproj)
# Set extent and plot map lines
ax.set_extent([-145., -70, 20., 60.], ccrs.PlateCarree())
ax.coastlines('50m', edgecolor='black', linewidth=0.75)
ax.add_feature(states_provinces, edgecolor='black', linewidth=0.5)
# Plot thickness with multiple colors
clevs = (np.arange(0, 5400, 60),
np.array([5400]),
np.arange(5460, 7000, 60))
colors = ('tab:blue', 'b', 'tab:red')
kw_clabels = {'fontsize': 11, 'inline': True, 'inline_spacing': 5, 'fmt': '%i',
'rightside_up': True, 'use_clabeltext': True}
for clevthick, color in zip(clevs, colors):
cs = ax.contour(lons, lats, thickness_1000_500, levels=clevthick, colors=color,
linewidths=1.0, linestyles='dashed', transform=dataproj)
plt.clabel(cs, **kw_clabels)
# Plot MSLP
clevmslp = np.arange(800., 1120., 4)
cs2 = ax.contour(lons, lats, mslp, clevmslp, colors='k', linewidths=1.25,
linestyles='solid', transform=dataproj)
plt.clabel(cs2, **kw_clabels)
# Use definition to plot H/L symbols
plot_maxmin_points(lons, lats, mslp, 'max', 50, symbol='H', color='b', transform=dataproj)
plot_maxmin_points(lons, lats, mslp, 'min', 25, symbol='L', color='r', transform=dataproj)
# Put on some titles
plt.title('MSLP (hPa) with Highs and Lows, 1000-500 hPa Thickness (m)', loc='left')
plt.title('VALID: {}'.format(vtime[0]), loc='right')
plt.show()
Total running time of the script: ( 0 minutes 3.132 seconds)